Research
AGMP, in partnership with ten Alzheimer Disease Research Centers across the US, is collecting fecal and blood samples from ADRC participants aged 60 and above using our at-home collection kits. We aim to recruit a racially diverse cohort of about 1000 participants, balanced between those diagnosed with Mild Cognitive Impairment, Alzheimer’s Disease, and controls. We will profile the samples using metagenomics and various metabolomics platforms across our consortium. Data generated will be analyzed and compared against existing datasets to elucidate links between the gut microbiome, metabolome, and Alzheimer’s disease pathogenesis.
Disease Stratification For Precision Medicine Approach To AD
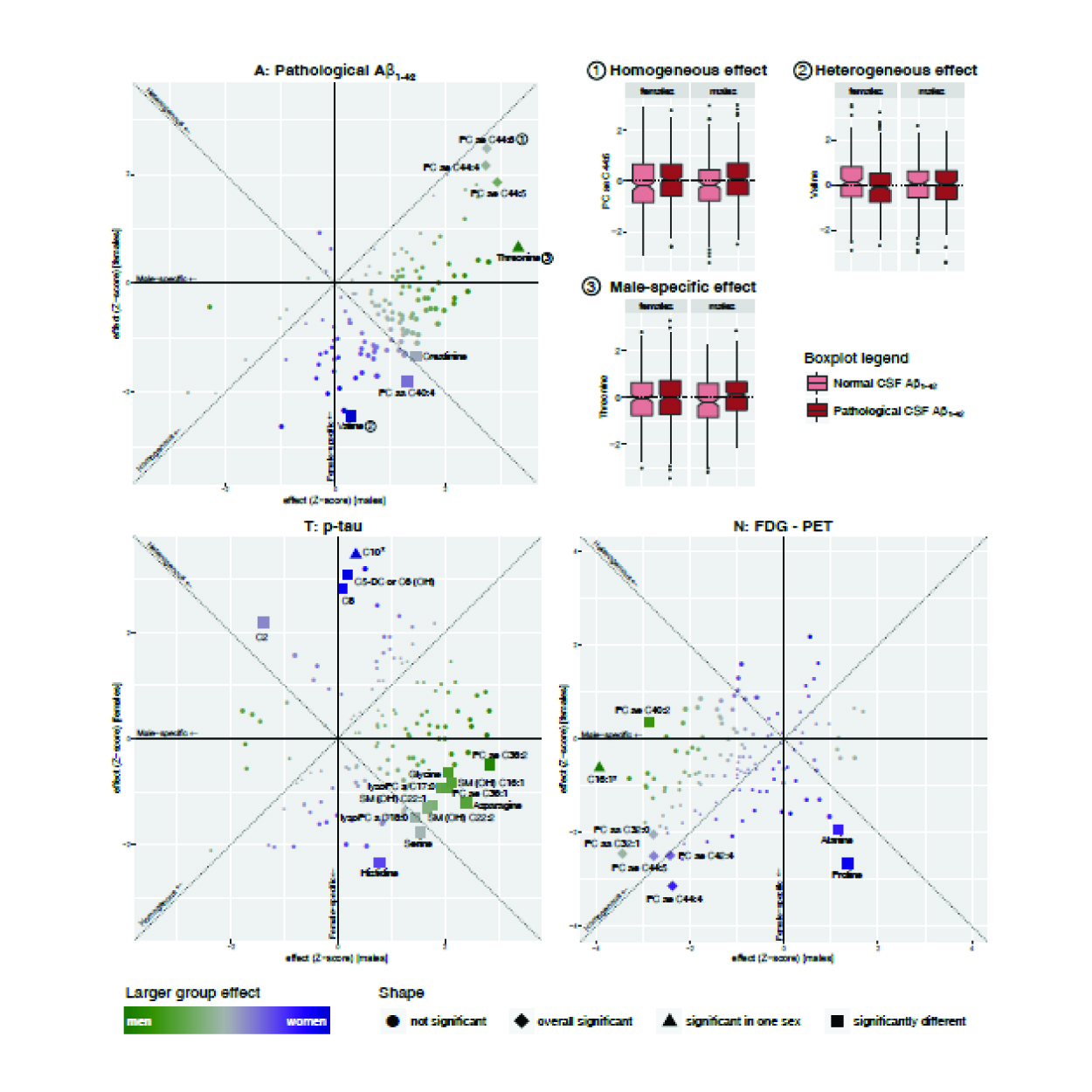
We systematically investigated group-specific metabolic alterations by conducting stratified association analyses of serum metabolites in individuals with measurements of AD biomarkers. Dissecting metabolic heterogeneity in AD pathogenesis can enable grading the biomedical relevance for specific pathways within specific subgroups, guiding the way to personalized medicine.
Gut-Liver-Brain Axis in AD
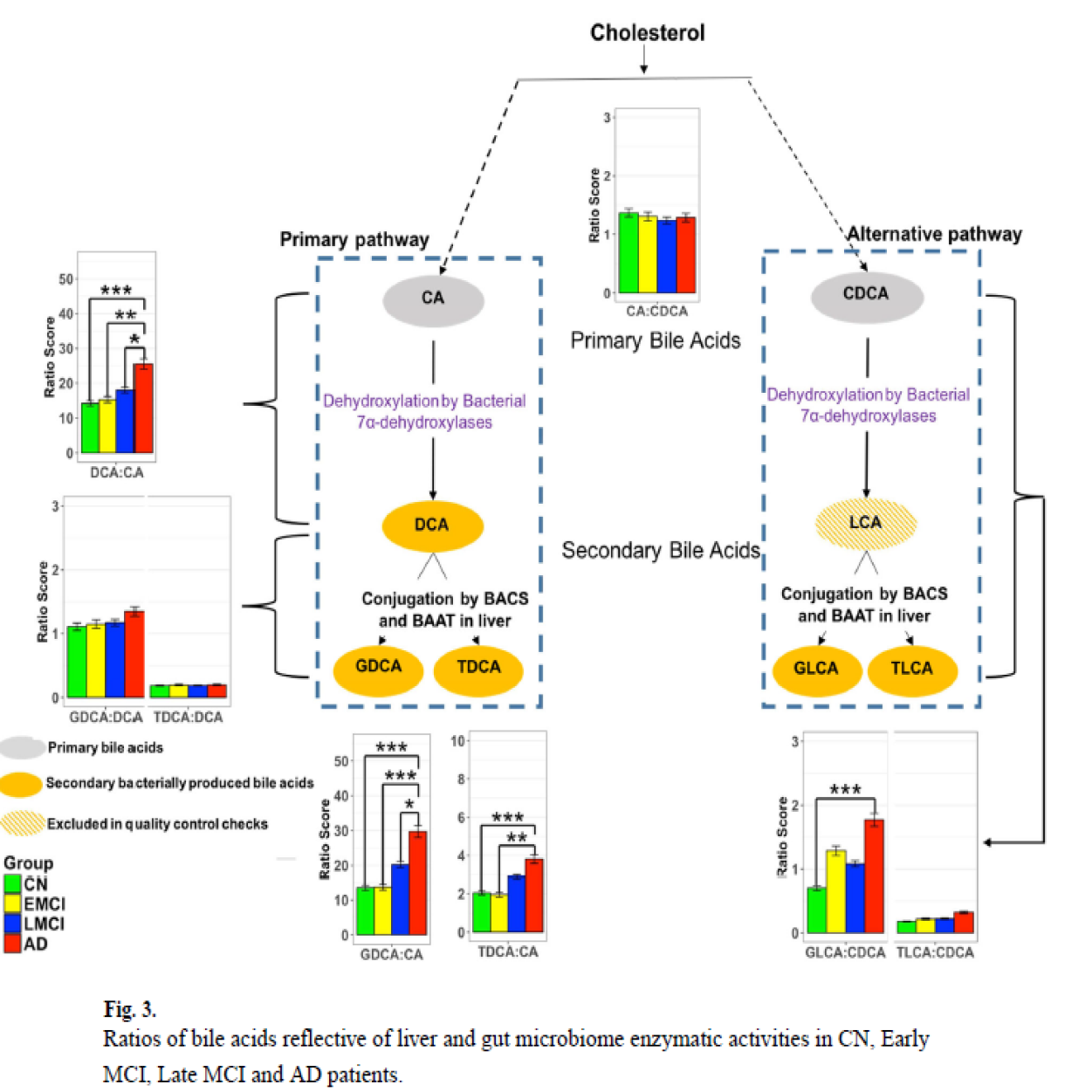
In two publications we showed an altered bile acid profile in AD was associated with cognitive decline as well as CSF amyloid-beta, p-tau, t-tau, and brain glucose metabolism providing further support for the role of BA pathways in AD pathophysiology.
Host-Microbiome Co-Metabolism
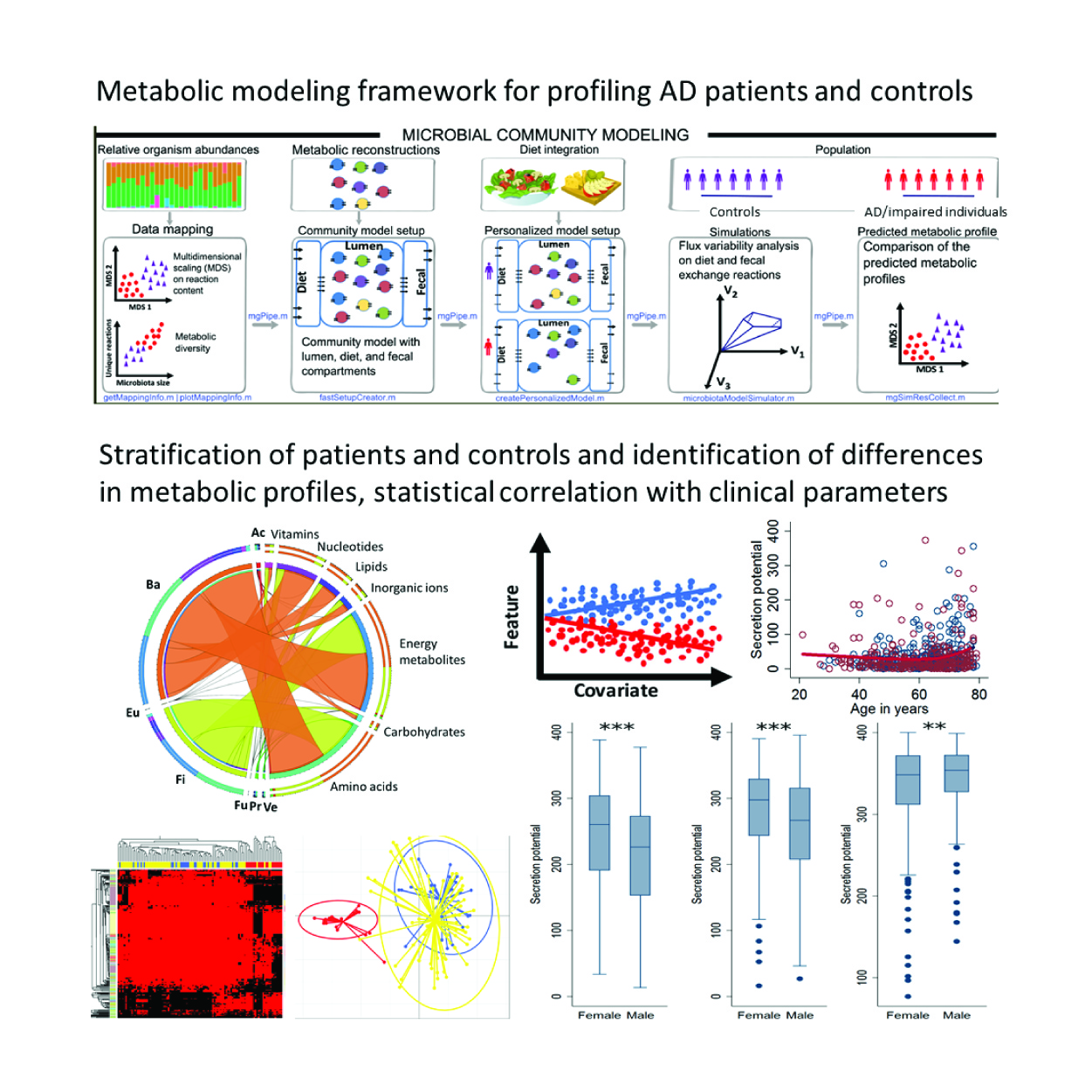
Using the COBRA approach, Dr. Ines Thiele and her team have generated the first physiologically resolved whole-body, gender-specific metabolic models (WBMs) based on extensive organ-specific proteomic and metabolomic data, as well as through literature curation.
To learn more visit:
https://www.thielelab.eu/research-projects

Molecular Atlas for AD
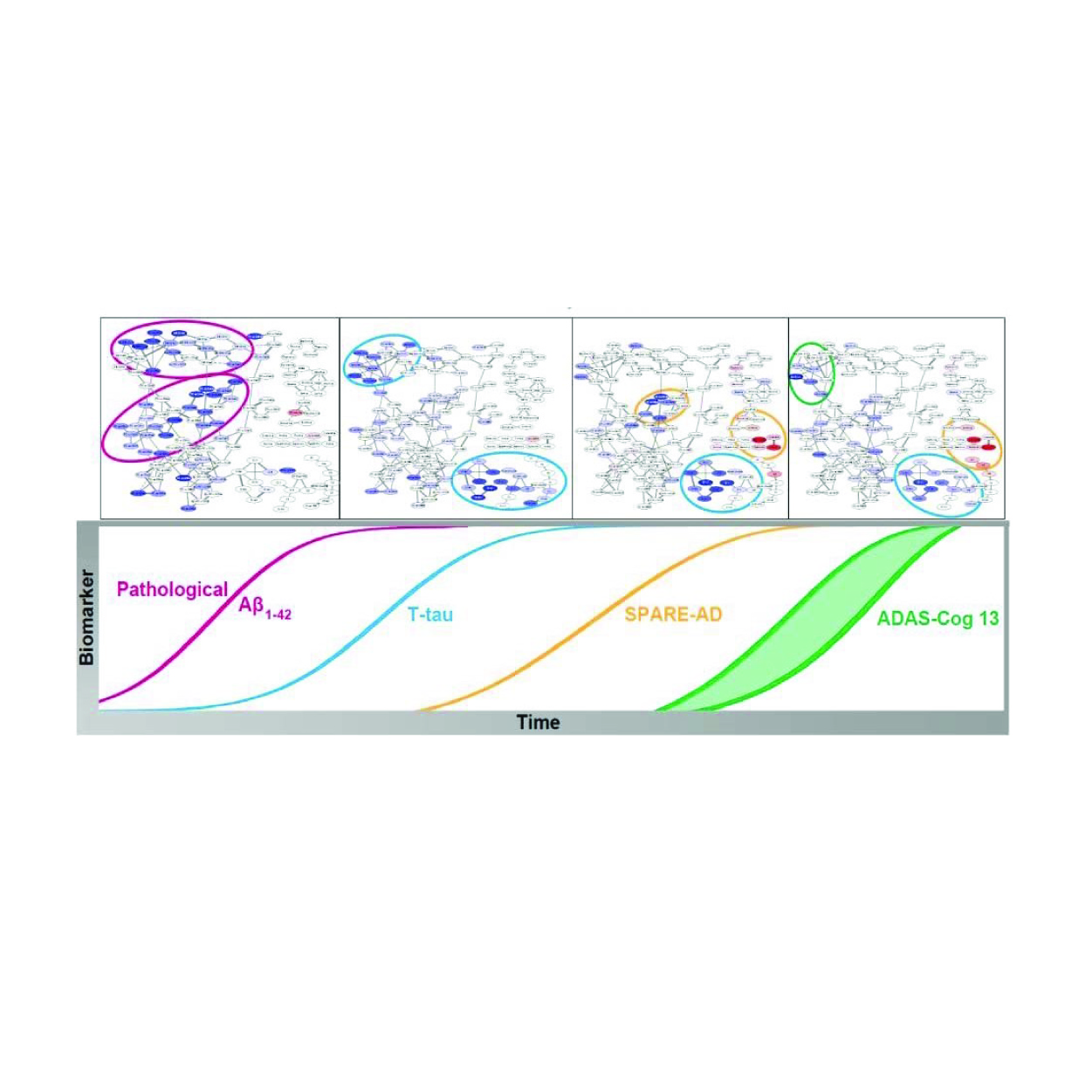
The AD atlas is a network-based data integration resource for investigating AD, its biomarkers, and associated endophenotypes in a multi-omics context. The database can be queried by entering one or more phenotypes, genes, proteins, or metabolites and provides an interactive interface to inspect generated networks, as well as enrichment tools for downstream analyses.
To learn more visit: https://adatlas.org/
Network-Based Approaches For Analayzing Metabolite Associations
We use metabolomics as a global biochemical approach to identify peripheral metabolic changes in AD patients and correlate them to AD biomarkers including cerebrospinal fluid pathology, brain imaging features, and cognitive performance.

Global Natural Products Social Molecular Networking
Global Natural Products Social Molecular Networking (GNPS) is a web-based mass spectrometry ecosystem that aims to be an open-access knowledge base for the community-wide organization and sharing of raw, processed, or identified tandem mass (MS/MS) spectrometry data. GNPS aids in identification and discovery throughout the entire life cycle of data; from initial data acquisition/analysis to post-publication.
To learn more visit: https://gnps.ucsd.edu/ProteoSAFe/static/gnps-splash.jsp
Quantitative Insights Into Microbial Ecology
QIIME 2™ is a next-generation microbiome bioinformatics platform that
is extensible, free, open-source, and community-developed.
For more information, visit: https://qiime2.org/

U.S. Pointer
The U.S. Study to Protect Brain Health Through Lifestyle Intervention to Reduce Risk (US POINTER) is a two-year clinical trial to evaluate whether lifestyle interventions that simultaneously target many risk factors protect cognitive function in older adults who are at increased risk for cognitive decline

Mind Diet Trial
Mediterranean-DASH Diet Intervention for Neurodegenerative Delay (MIND) Trial to Prevent Alzheimer’s is a three-year research study that compares two weight-loss diets and their effects on brain health and cognitive decline.

BEAT-AD Clinical Trial
Brain Energy for Amyloid Transformation in Alzheimer’s Disease Study (BEAT-AD) compares the effects of a ketogenic low-carbohydrate diet and a low-fat diet in adults with mild cognitive impairment. The data collected will help determine whether diet interventions induce changes in cognition, cerebral blood flow, and levels of certain proteins and hormones.
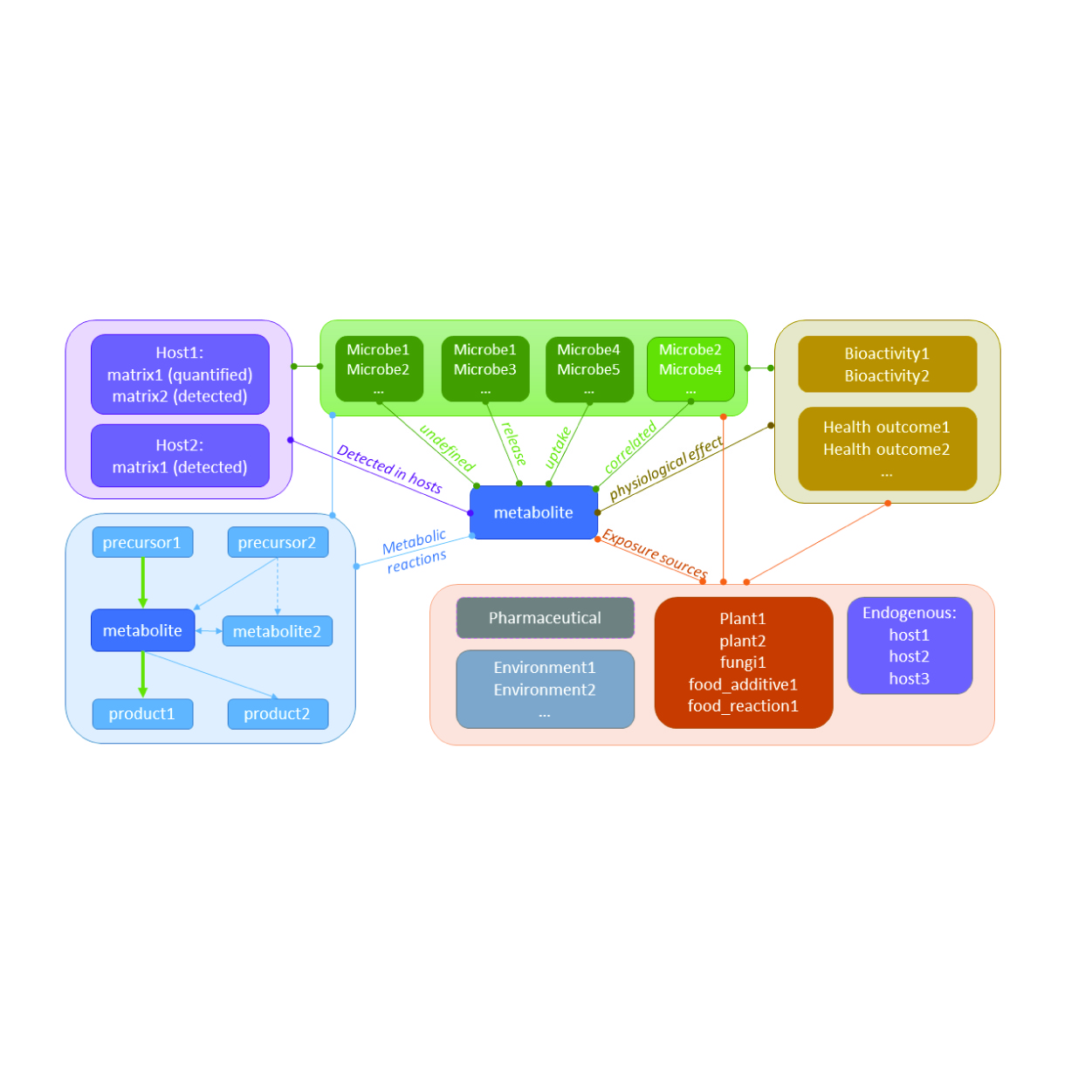
Microbial Metabolite Database
Through our partnership with Leiden University and the greater metabolomics community, we are working to establish a comprehensive database for microbial metabolites. Interconnected modules in the database will allow for dynamic browsing and search combinations.
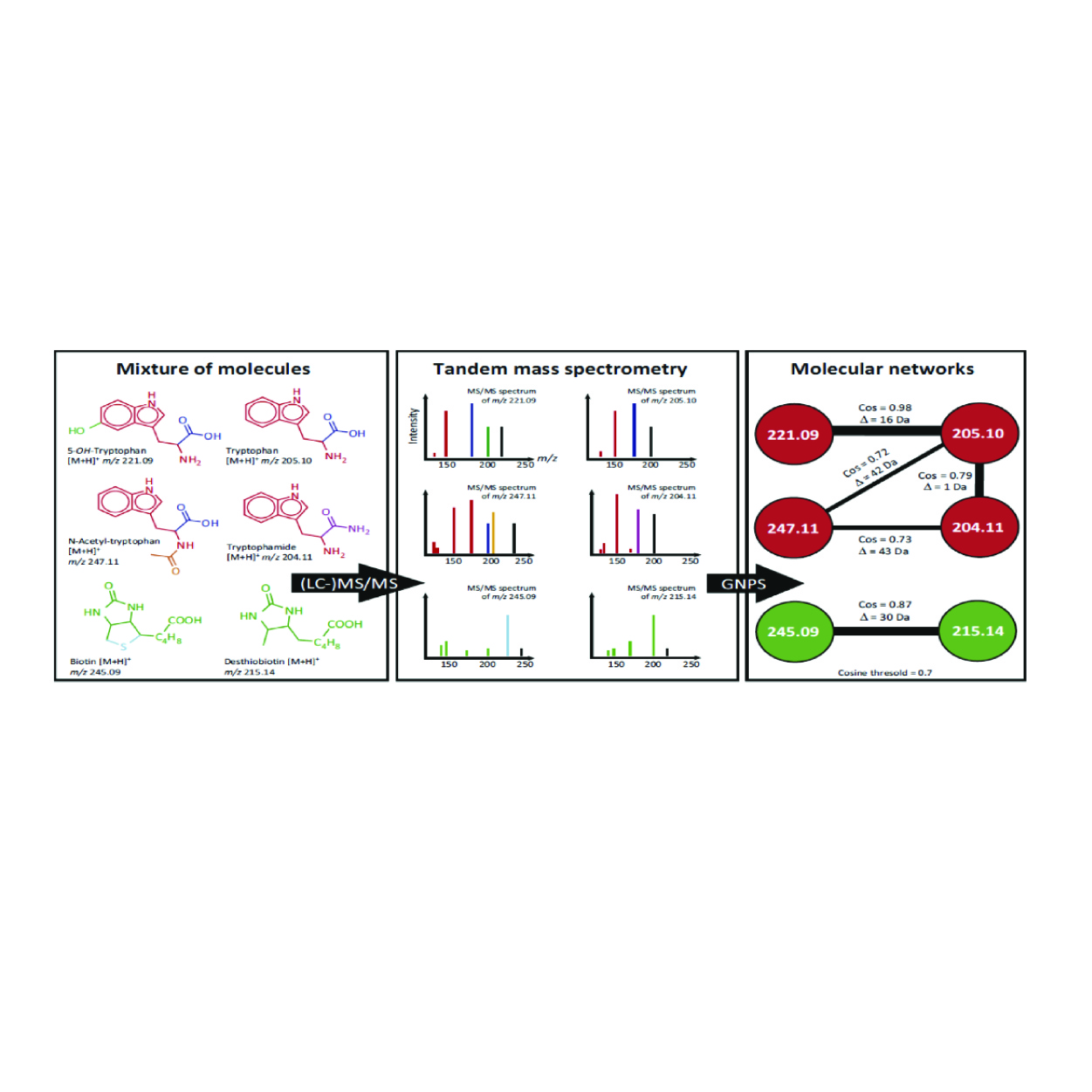
Molecular Networking
Molecular networking is a strategy that first merges all identical spectra and then creates a map of MS/MS similarity from which chemical similarity can be inferred. Molecular networking allows the discovery of molecules that are not identical but are related to known molecules.
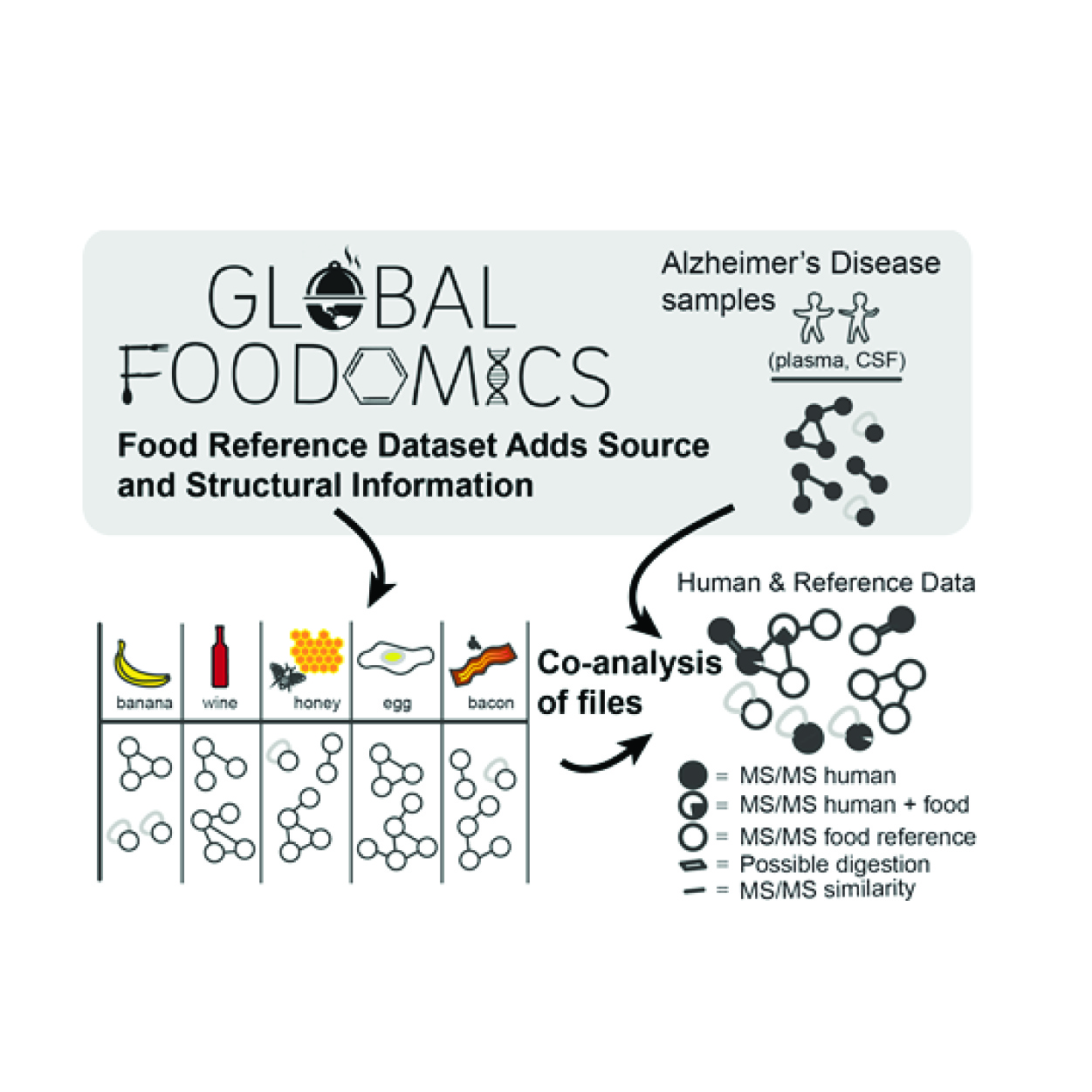
Global Food Omics
The Global Food Omics initiative was launched at UC San Diego in collaboration with the American Gut Project, to learn more about bacteria in foods and the small molecules they produce.
To learn more visit: Global Food Omics

Qiita
Qiita (canonically pronounced cheetah) is an entirely open-source microbial study management platform. It allows users to keep track of multiple studies with multiple ‘omics data. Additionally, Qiita is capable of supporting multiple analytical pipelines through a 3rd-party plugin system, allowing the user to have a single entry point for all of their analyses.
Qiita provides database and computes resources to the global community, alleviating the technical burdens that are typically limiting for researchers studying microbial ecology (e.g. familiarity with the command line or access to compute power).
Qiita’s platform allows for quick reanalysis of the datasets that have been deposited using the latest analytical technologies. This means that Qiita’s internal datasets are living data that is periodically re-annotated according to current best practices.
For more information about how to use Qiita, visit the documentation: https://qiita.ucsd.edu/static/doc/html/index.html
